-
Posts
17 -
Joined
-
Last visited
Content Type
Profiles
Forums
Downloads
Blogs
Posts posted by Sehaj
-
-
Image ID TRDR DDR Obs Time (Earth) Season in SH MY Ls
MSV0003211D_01 MROCR_2109 MROCR_1007 26-08-2014 spring 32 184.6
@Ray Arvidson please find the details
-
Hello community,
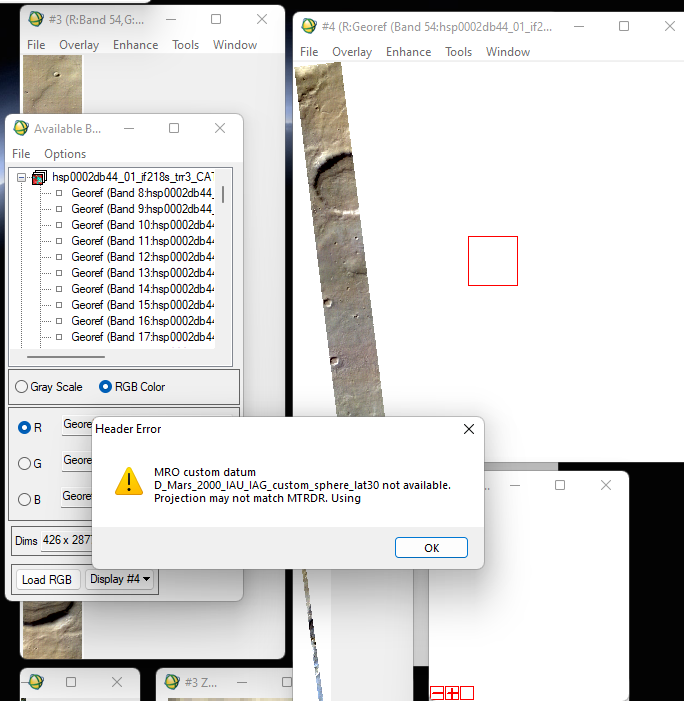
While working with MSV, HSV crism ids of a region near the south pole, am finding strange results after processing CRISM cubes. These crism observations belong to the same period of the year (between Ls 180-270). While some crism images after atm correction and georeferencing seems fine, other show different colours and strange patterns (diagonal dark streak lines) and horizontal layers of diff colours (rightmost strip over the study region).
While same procedure for processing the cubes has been applied, why some results are showing strange patterns. Does anyone have better understanding? (See attachment)
-
I'll like to point I ve been processing some CRISM cubes, and have realized that the processing (particularly, projecting the cube) time is much longer in ENVI 5.6 in comparison to ENVI 5.0.
For instance, an MSV took 10-12 mins to georeference in latest version, while it took 2-3 mins in the former.
Can anyone from the developers team verify and tell why this is happening so?
-
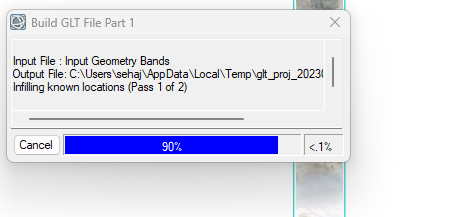
Hey,
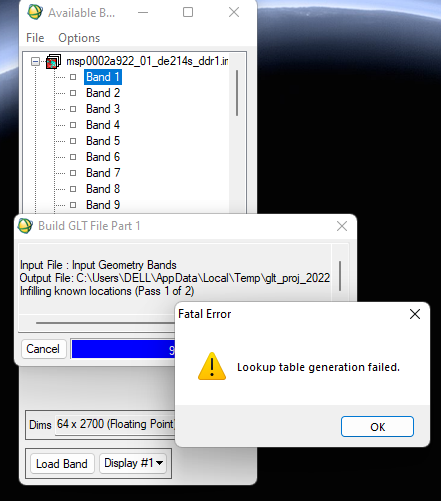
I've been using CAT 7.4 for a long time now. I don't know what has gone wrong. A single CRISM cube is taking approx 10 mins to build GLT file ( see screenshot) while projecting, which earlier used to take seconds.
I have ENVI 5.6 installed on my i9 processor PC with 64 GB RAM. So, I am sure it's not about hardware. Please, anyone, help as I have a plethora of CRISM cubes to process.
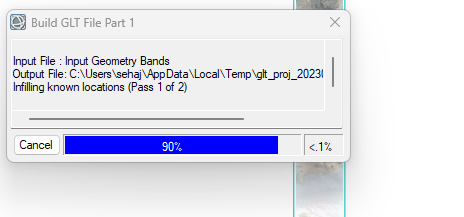
Thanks in anticipation!
-
Hello Feng,
Remember we did a zoom meeting to solve the GLT lookup table error and header error. We managed to fix the fatal error but the header error persisted.
I am not getting any kind of errors, though, but projecting a single cube image is taking a lot of time (10mins approx). I have a plethora of images to process, and need to fix this issue. At the time of zoom call, this problem came and you resolved the problem.
Can you please help me in getting over this?
Thanks,
Sehajpal Singh
-
Yes, seen now...Thanks!
-
How can we apply correction for aerosols on CRISM data?
Is there any option for the same in CAT toolkit?
-
Does CAT toolkit take care of aerosol correction? How can we apply aerosol correction to CRISM cube?
-
Okay..Many thanks!
-
Thanks for this information.
I hope by flattening the sensor space data, you mean to say dividing the spectra by the spectra of neutral region in my image?
-
While using CAT toolkit, we go for cleaning (destriping and despiking for IR, and destriping for VNIR) the data. In Data Filtering, there are two alternatives:
1. CIRRUS>Clean spectral cube
2. MRO CRISM Remove Stripes
I see generally, CIRRUS method is preferred. What is the science behind cleaning by two methods. And which method should be optimum for me?
What if, I do not go for data filtering and simply georeference the image after photometric and atmospheric corrections?
Thanks in advance.
-
That'll be so nice of you.
I ll send you the details and you finalize the slot as per your convenience.
-
-
Thanks Feng. I am using ENVI 5.6 (IDL 8.8). I have been using ENVI classic for processing via CAT toolkit and performing other geoprocessing operations in standard version.
-
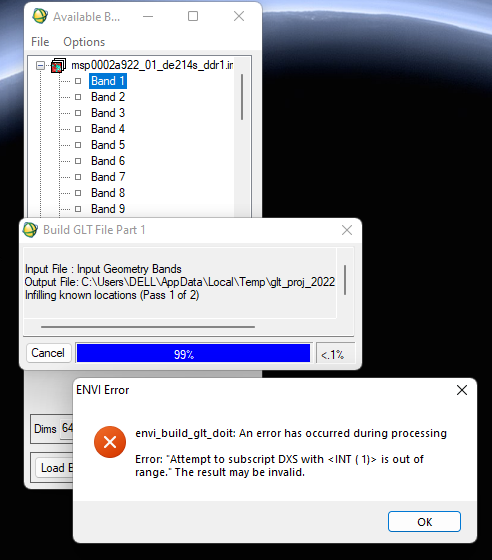
Thanks for your quick response. No, I destriped before going for projecting.
Please let me correct that after atmospheric and photometric correction, am applying destriping, followed by projection (As the processed filename suggests: hsp..._corr_destripe_p.img) . I am afraid if HSP data has some problem, because I did not face any issues while processing MSP dataset.
However, I keep on getting the following error while projecting with respective ddr files. Although, the final image is projected and shows coordinates when we move cursor across it.
Can you tell what is the problem?
-
Hello CRISM Team!
I have been using CRISM data for around 2 years and I appreciate the team for providing useful workshops and TRR-V3 data. I am wondering no new workshops have came up for using CRISM data.
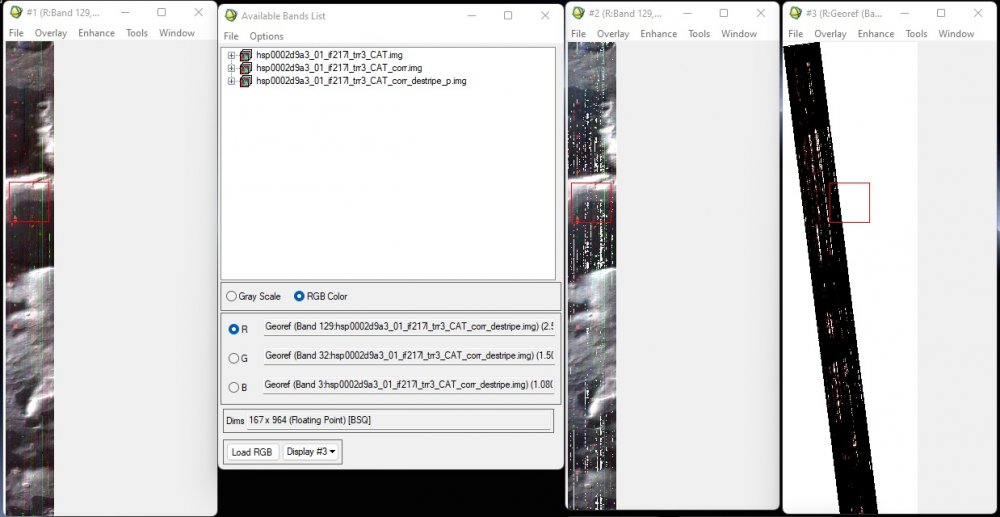
Recently, I have processed MSP and HSP images (TRDR) of Hellas Planitia region. In HSP dataset, I am getting a lot of bad bands. Though ATP and PHT corrections ignore some bad bands, but after applying these corrections, hsp0002d9a3 is getting worse (Display 2 in screenshot) as there might be more bad bands. Projecting the cube and applying destriping (Display 3 in screenshot) gives unusual results.
Similar issues are being faced while processing some other files.
Do, I need to spectrally subset good bands individually from each of the files? I am afraid there are lot of bands and many files, so this would be a a bit difficult job.
Or else, can I safely ignore this data and try with different observations.
Thanks in anticipation!
- Sehajpal Singh (Doctoral candidate at IIT Bombay)


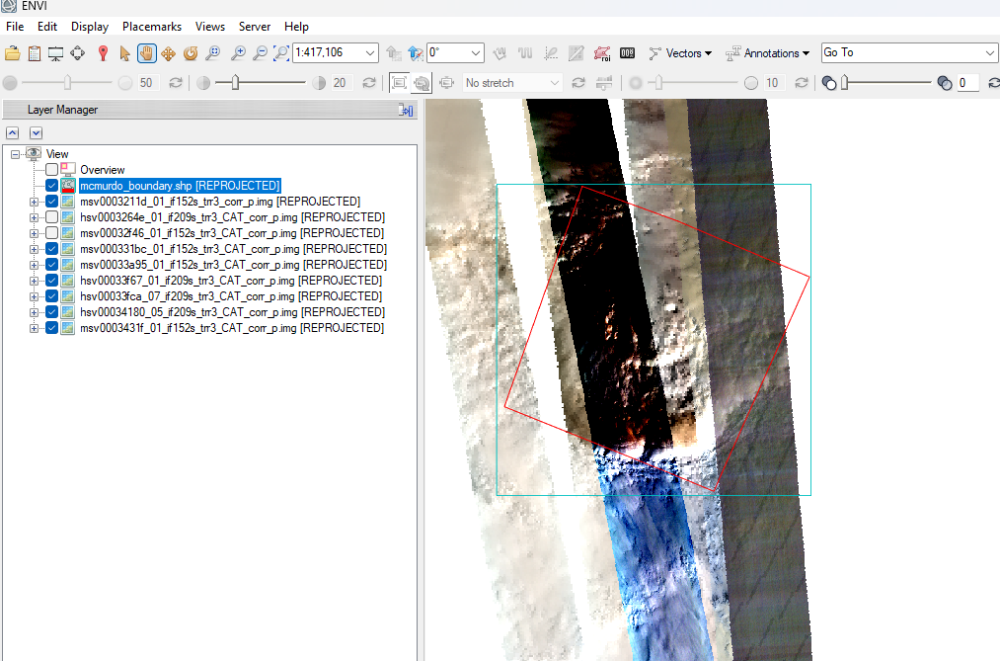
CRISM observations anomalies over South Polar Region
in For data users
Posted
Okay thanks. I will look into it