
Subhabose
-
Posts
8 -
Joined
-
Last visited
Content Type
Profiles
Forums
Downloads
Blogs
Posts posted by Subhabose
-
-
June,
I have used the GLT procedure in ENVI to georeference M3 images. The process is working now.
Thanks!
-
Hi,
I have attached the map_proj.txt, datum.txt and the ellipse.txt files with the updated strings. The image I am using is M3G20081229T022350_V01_RFL.
-
Hi,
The product ID is M3G20081229T022350. The projection used was Lunar Sinusoidal. I tried to follow the steps prescribed by you for assigning map information to PDS images in ENVI.
-
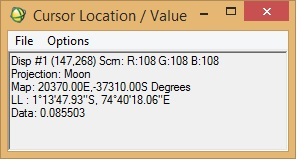
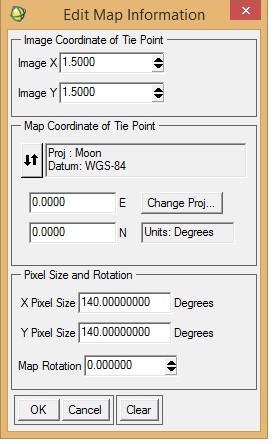
I want to georeference an M3 image (obtained via PDS) in ENVI. I was following "Assigning Map Information to PDS Images in ENVI", posted by Dr June Wang. I was able to perform most of the steps. Yet when the process is complete, it seems that the georeferencing is not correct. I have attached a couple of screenshots from the M3 image I was working with.
To check the accuracy of georeferencing, I cross-checked the location from Google Moon and found that there are a lot of discrepancies between the 'georeferenced' M3 image and Google Moon. In the 'Edit Map Information' window, I entered X-pixel and Y-pixel sizes as 140. Please let me know if this is correct. When I cross-checked with Google Moon, the top of the image should have latitude as north and the bottom the image as south (also verified from LOC file of the M3 image). Please help.
-
-
On 11/6/2017 at 9:35 PM, Ray Arvidson said:
Greetings:
You can use both the new ENVI5 series interfaces or the Classic ENVI interface to a resample spectral data set to a new set of center wavelengths and, optionally, band passes. You will find the spectral resampling procedure under the toolbox on the right hand side of the ENVI5 series interface, and under the spectral heading on the pull down Classic menu. Next select an input data file (it needs to have wavelengths in the header record) that you designate as the new wavelengths for the spectrally resampled data. The dialog box then asks you for the output file that will be resampled. If there are band passes (like Gaussians) in the input data file header these will also be used in resampling the output data set. I just tried it with an OMEGA L data set as the input and one of the ENVI JHU spectral libraries as an example and it worked. It resampled the JHU data to the OMEGA wavelengths and made a new JHU spectral library. Also works with spectral cubes as input and output data sets, etc. Let me know if you still have some problems and we can talk on the phone.
Ray Arvidson
PDS Geosciences Ndoe Manager.
11/6/17
Hello!
Thank you very much for your reply!
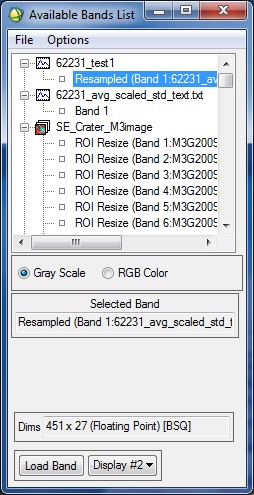
I followed the steps mentioned using ENVI Classic and am sharing some of the results with you.
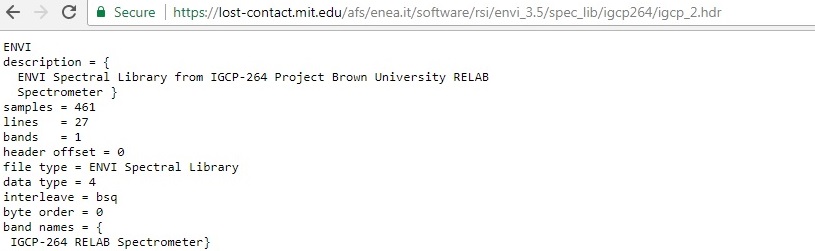
First, I chose the 62231 ASCII file and converted it to a spectral library using the RELAB ENVI header file information, which I found here (https://lost-contact.mit.edu/afs/enea.it/software/rsi/envi_3.5/spec_lib/igcp264/igcp_2.hdr).
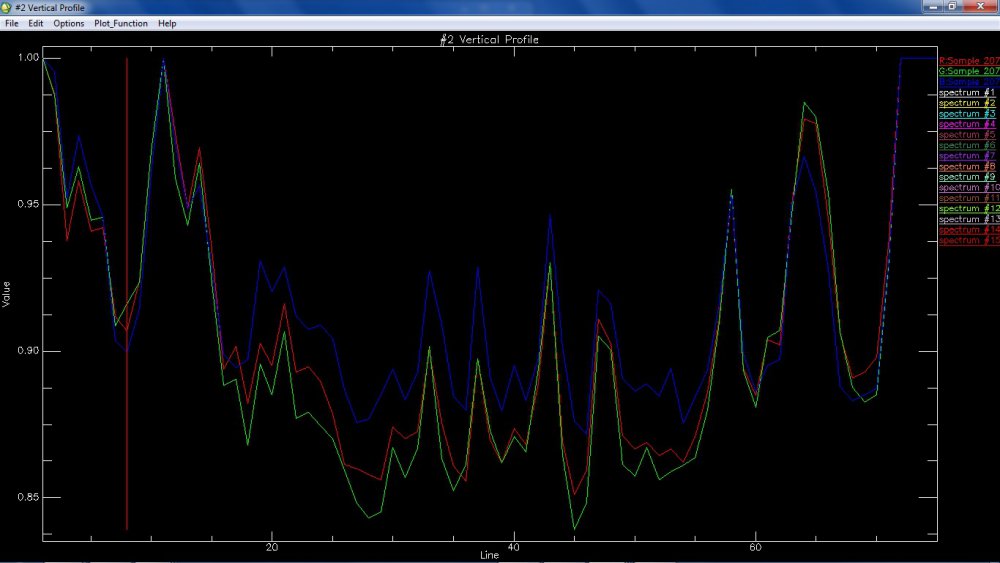
Once that was done, I tried to match the same with a small region that I was able to subset from one of the M3 (Level 2) images. Unfortunately, I am still unable to match the two spectra.
Please let me know where I am going wrong.
For your convenience, I am attaching some screenshots.
-
Hi!
I want to compare spectra generated from M3 with RELAB library spectra (62231, Apollo 16 landing site soil sample) using ENVI. One specific issue is, spectral library from RELAB (62231) has 451 bands, whereas M3 has 86 spectral bands. This is apparently creating a mismatch and is not allowing me to compare the two spectra.
Please help!
Reading Clementine images
in For data users
Posted
I wish to utilize selected portions of Clementine NIR data. I downloaded a few files from the Lunar ODE.
One of the downloaded folders was 'CL_0064', which further led to lun276 and lun277 folders and many subsequent folders. The final file had a (.276) extension, which I am not able to open in ENVI. Please let me know the correct procedure to open such files in ENVI or ArcMap.
I also downloaded 'clemdcmp' and 'climdisp' from https://pdsimage2.wr.usgs.gov/archive/clem1-l_e_y-a_b_u_h_l_n-2-edr-v1.0/cl_0064/software/ . However, I am not able to install either of the software. Please help!